With the growth in genomic data availability, there is a need for tools that scale better.
I have been involved with the development of tools that enable scalable and reproducible simulations,
within stdpopsim and tskit.
More recently, I have been developing a deep learning method for population genetic inference that takes tree sequences, a scalable format for population genomic data, with Andrew Kern and Nate Pope
Efficient ancestry and mutation simulation with msprime 1.0Franz Baumdicker, Gertjan Bisschop, Daniel Goldstein, Graham Gower, Aaron P Ragsdale, Georgia Tsambos, Sha Zhu, Bjarki Eldon, E Castedo Ellerman, Jared G Galloway, Ariella L Gladstein, Gregor Gorjanc, Bing Guo, Ben Jeffery, Warren W Kretzschumar, Konrad Lohse, Michael Matschiner, Dominic Nelson, Nathaniel S Pope, Consuelo D Quinto-Cortés, Murillo F Rodrigues, Kumar Saunack, Thibaut Sellinger, Kevin Thornton, Hugo Van Kemenade, Anthony W Wohns, Yan Wong, Simon Gravel, Andrew D Kern, Jere Koskela, Peter L Ralph, Jerome Kelleher
Expanding the stdpopsim species catalog, and lessons learned for realistic genome simulationsM Elise Lauterbur, Maria Izabel a Cavassim, Ariella L Gladstein, Graham Gower, Nathaniel S Pope, Georgia Tsambos, Jeffrey Adrion, Saurabh Belsare, Arjun Biddanda, Victoria Caudill, Jean Cury, Ignacio Echevarria, Benjamin C Haller, Ahmed R Hasan, Xin Huang, Leonardo Nicola Martin Iasi, Ekaterina Noskova, Jana Obsteter, Vitor Antonio Correa Pavinato, Alice Pearson, David Peede, Manolo F Perez, Murillo F Rodrigues, Chris CR Smith, Jeffrey P Spence, Anastasia Teterina, Silas Tittes, Per Unneberg, Juan Manuel Vazquez, Ryan K Waples, Anthony Wilder Wohns, Yan Wong, Franz Baumdicker, Reed a Cartwright, Gregor Gorjanc, Ryan N Gutenkunst, Jerome Kelleher, Andrew D Kern, Aaron P Ragsdale, Peter L Ralph, Daniel R Schrider, Ilan Gronau
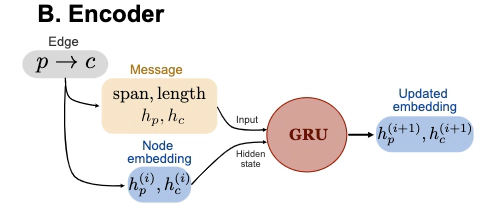 Encoder of a message-passing deep learning network.
Encoder of a message-passing deep learning network.